Kaggle Project: Categorical Feature Engineering
About
This dataset is obtained from kaggle Credit Cards Categorical Feature Encoding Challenge II that can be found here: https://www.kaggle.com/c/cat-in-the-dat-ii
Content
Hello all, in this dataset we are given a bunch of categorical features like the previous one; except that this one has missing values. So we are going to approach the problem in 3 steps. First one is to fill these missing values,then we’re going to encode them. Finally, we will fit a model. In this dataset, I chose to use Logistic Regression with regularization. We have binary, nominal and ordinal features. Lets begin.
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
train = pd.read_csv("../input/cat-in-the-dat-ii/train.csv")
test = pd.read_csv("../input/cat-in-the-dat-ii/test.csv")
# Subset
target = train['target']
train_id = train['id']
test_id = test['id']
train.drop(['id'], axis=1, inplace=True)
test.drop('id', axis=1, inplace=True)
#Null values
null_df = pd.DataFrame({'Percentile':train.isnull().sum()/len(train), 'Count':train.isnull().sum()})
print(null_df)
All variables has missing values around 3%.
my_df = pd.concat([train, test])
my_df.isnull().sum()
- bin_0 29795
- bin_1 30041
- bin_3 29965
- bin_4 29998
- day 29977
- month 29972
- nom_0 30314
- nom_1 30103
- nom_2 30214
- nom_3 30297
- nom_4 30028
- nom_5 29690
- nom_6 30143
- nom_7 30006
- nom_8 29711
- nom_9 30133
- ord_0 30181
- ord_1 30208
- ord_2 30180
- ord_3 29969
- ord_4 29863
- ord_5 29760
- target 400000
- dtype: int64
Preprocessing
Start with binary ones.
my_df["bin_3"] = my_df["bin_3"].apply(lambda x: 1 if x=='T' else (0 if x=='F' else None))
my_df["bin_4"] = my_df["bin_4"].apply(lambda x: 1 if x=='Y' else (0 if x=='N' else None))
In nominal and ordinal ones, first we have to convert to string by .astype. But when we do this, NaN values are converted to string “nan”. So we will write them None afterwards.
for enc in ["nom_0","nom_1","nom_2","nom_3","nom_4","day","month","ord_3"]:#,"ord_4","ord_5"]:
my_df[enc] = my_df[enc].astype("str")
my_df[enc] = my_df[enc].apply(lambda x: x.lower())
my_df[enc] = my_df[enc].apply(lambda x: None if x=='nan' else x)
for enc in ["ord_4","ord_5"]:
my_df[enc] = my_df[enc].astype("str")
my_df[enc] = my_df[enc].apply(lambda x: None if x=='nan' else x)
ord_1 and ord_2 can be encoded like the following. They include ordering such as Warm>Cold>Freezing etc.
my_df["ord_1"] = my_df["ord_1"].apply(lambda x: 1 if x=='Novice' else (2 if x=='Contributor' else (3 if x=='Expert' else (4 if x=='Master' else (5 if x=='Grandmaster' else None)))))
my_df["ord_2"] = my_df["ord_2"].apply(lambda x: 1 if x=='Freezing' else (2 if x=='Cold' else (3 if x=='Warm' else (4 if x=='Hot' else (5 if x=='Boiling Hot' else (6 if x=='Lava Hot' else None))))))
Now here comes the important part. As Bojan mentioned here , the most important features are the weird ones, which are nom_5, nom_6, nom_7, nom_8 and nom_9. I filled the missing values of these with mode and used these features to impute the missing in other variables with similar values. The reason that I filled with mode is, when we use pandas’ group by function, if one of the grouped variables are missing, then it returns NaN. So they have to be complete.
for col in ["nom_5","nom_6","nom_7","nom_8","nom_9"]:
mode = my_df[col].mode()[0]
my_df[col] = my_df[col].astype(str)
my_df[col] = my_df[col].apply(lambda x: mode if x=='nan' else x)
Get the other variables list. And fill wtih similar values.
columns = list(my_df.columns)
for col in ["target","nom_5","nom_6","nom_7","nom_8","nom_9"]:
columns.remove(col)
I used nom_7 to group only since it looks like the most important variable according to the feature importances from the link I mentioned above. Check this pic:
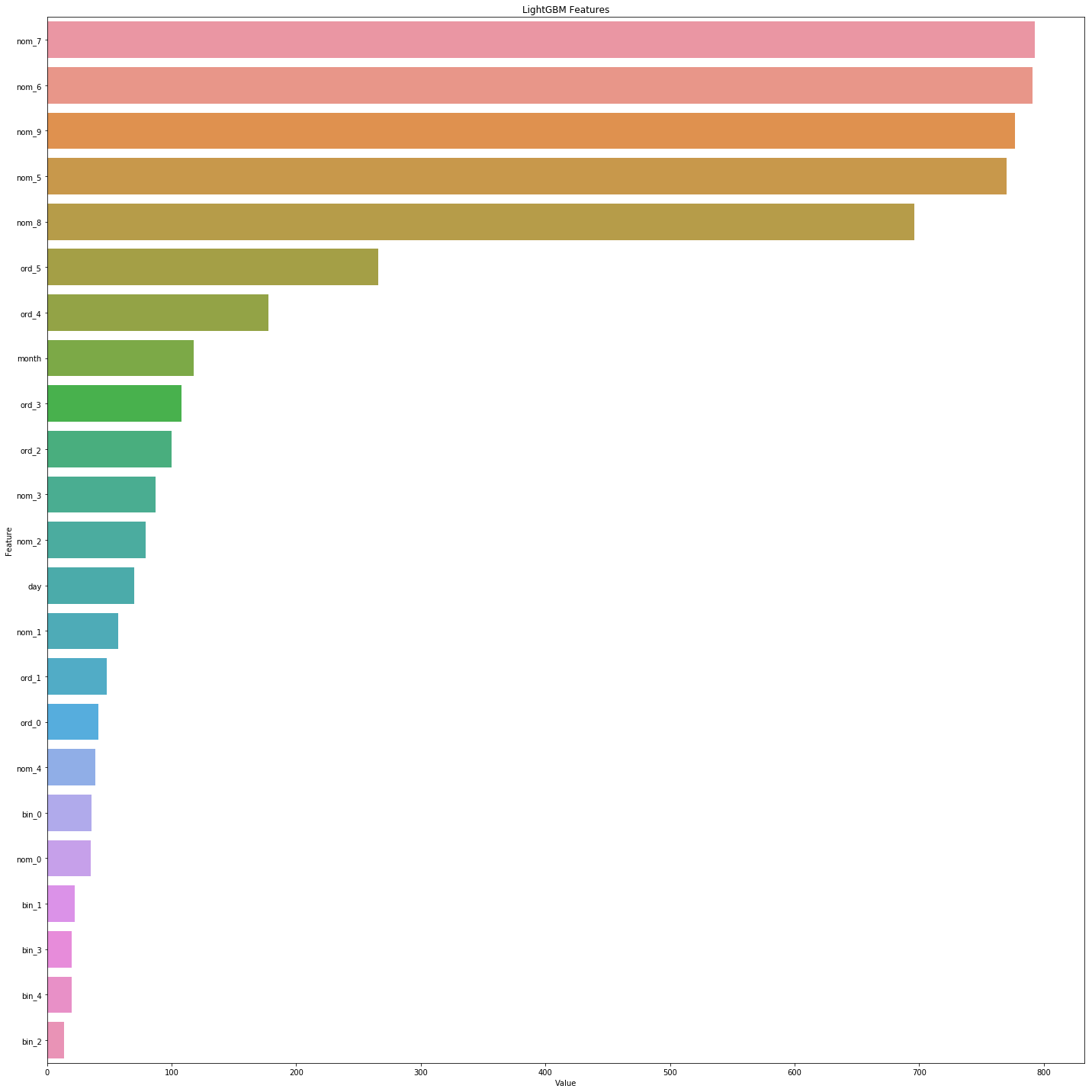
The reason we didn’t use more variables to group by is, grouping with high-dimensional features is not efficient. Sometimes it creates bias and result in overfitting. This way we are kinda safe. After grouping, we will fill with the mode value of that group.
for col in columns:
my_df[col] = my_df.groupby(["nom_7"])[col].transform(lambda x: x.fillna(x.mode()[0]))
Now check the missing values.
my_df.isnull().sum()
- bin_0 0
- bin_1 0
- bin_2 0
- bin_3 0
- bin_4 0
- day 0
- month 0
- nom_0 0
- nom_1 0
- nom_2 0
- nom_3 0
- nom_4 0
- nom_5 0
- nom_6 0
- nom_7 0
- nom_8 0
- nom_9 0
- ord_0 0
- ord_1 0
- ord_2 0
- ord_3 0
- ord_4 0
- ord_5 0
- target 400000
No missing values left except for test set’s target. Now, lets encode the remaining variables. I will convert ordinal ones with ordinal encoder. It will assign 1,2,3… according to alphabetic order of string variables. Note that ord_4 and ord_5 include both upper and lower case letters. They might include information. So lets use ordinal encoder on them too.
from sklearn.preprocessing import OrdinalEncoder
oencoder = OrdinalEncoder(dtype=np.int16)
for enc in ["ord_3","ord_4","ord_5"]:
my_df[enc] = oencoder.fit_transform(np.array(my_df[enc]).reshape(-1,1))
We can get dummies for the rest of the categoric variables. For high dimensional ones, we will only create dummies for 2 of them.
for category in ["nom_5","nom_6","nom_7","nom_8","nom_9"]:
print("{} has {} unique values".format(category,len(np.unique(my_df[category]))))
- nom_5 has 1220 unique values
- nom_6 has 1520 unique values
- nom_7 has 222 unique values
- nom_8 has 222 unique values
- nom_9 has 2218 unique values
nom_7 and nom_8 has lower dimension. We can use pandas’ dummy encoder on them. If we use it on all, it will increase the width of our train set too much and it will create bugs/errors on direct model fitting. This way still performs well. We will use another encoding for high-dim variables later.
for enc in ["nom_0","nom_1","nom_2","nom_3","nom_4","day","month","nom_7","nom_8"]:
enc1 = pd.get_dummies(my_df[enc], prefix=enc)
my_df.drop(columns=enc, inplace=True)
my_df = pd.concat([my_df,enc1], axis=1)
Scale the ordinal and binary variables (-1,1). This can help linear models.
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler((-1,1))
for feat in ["ord_0","ord_1","ord_2","ord_3","ord_4","ord_5"]:
my_df[feat] = scaler.fit_transform(np.array(my_df[feat]).reshape(-1,1))
for feat in ["bin_0","bin_1","bin_2","bin_3","bin_4"]:
my_df[feat] = scaler.fit_transform(np.array(my_df[feat]).reshape(-1,1))
Create train and test.
test = my_df[my_df["target"].isnull()]
test.drop(columns='target', inplace=True)
train = my_df[my_df["target"].isnull()==False]
target = train["target"]
Before dropping target column from train set, I will use LeaveOneEncoder on high dimensional nominal variables.
from category_encoders import LeaveOneOutEncoder
leaveOneOut_encoder = LeaveOneOutEncoder()
for nom in ["nom_5","nom_6","nom_9"]:
train[nom] = leaveOneOut_encoder.fit_transform(train[nom], train["target"])
test[nom] = leaveOneOut_encoder.transform(test[nom])
Now lets drop the target and reset indexes.
train.drop(columns='target', inplace=True)
train.reset_index(drop=True, inplace=True)
test.reset_index(drop=True, inplace=True)
target.reset_index(drop=True, inplace=True)
Modelling
Import the libraries we gonna need.
from sklearn.model_selection import KFold
from sklearn.metrics import roc_auc_score as auc
from sklearn.linear_model import LogisticRegression
def run_cv_model(train, test, target, model_fn, params={}, eval_fn=None, label='model'):
kf = KFold(n_splits=5)
fold_splits = kf.split(train, target)
cv_scores = []
pred_full_test = 0
pred_train = np.zeros((train.shape[0]))
i = 1
for dev_index, val_index in fold_splits:
print('Started ' + label + ' fold ' + str(i) + '/5')
dev_X, val_X = train.loc[dev_index], train.loc[val_index]
dev_y, val_y = target.loc[dev_index], target.loc[val_index]
params2 = params.copy()
pred_val_y, pred_test_y = model_fn(dev_X, dev_y, val_X, val_y, test, params2)
pred_full_test = pred_full_test + pred_test_y
pred_train[val_index] = pred_val_y
if eval_fn is not None:
cv_score = eval_fn(val_y, pred_val_y)
cv_scores.append(cv_score)
print(label + ' cv score {}: {}'.format(i, cv_score))
i += 1
print('{} cv scores : {}'.format(label, cv_scores))
print('{} cv mean score : {}'.format(label, np.mean(cv_scores)))
print('{} cv std score : {}'.format(label, np.std(cv_scores)))
pred_full_test = pred_full_test / 5.0
results = {'label': label,
'train': pred_train, 'test': pred_full_test,
'cv': cv_scores}
return results
def runLR(train_X, train_y, test_X, test_y, test_X2, params):
print('Train LR')
model = LogisticRegression(**params)
model.fit(train_X, train_y)
print('Predict 1/2')
pred_test_y = model.predict_proba(test_X)[:, 1]
print('Predict 2/2')
pred_test_y2 = model.predict_proba(test_X2)[:, 1]
return pred_test_y, pred_test_y2
Set inverse regularization parameter C=0.1 and initialize.
lr_params = {'solver': 'lbfgs', 'C':0.1, 'max_iter':500}
results = run_cv_model(train, test, target, runLR, lr_params, auc, 'lr')
- lr cv score 5: 0.7847225367977033
- lr cv scores : [0.7844204265213018, 0.7848292810796071, 0.7853701511296699, 0.7853865447776116, 0.7847225367977033]
- lr cv mean score : 0.7849457880611789
- lr cv std score : 0.0003778280688249727
Our mean CV value is 78,5. For this much feature engineering, it is sufficient I suppose :) Let’s submit the results.
submission = pd.DataFrame({'id': test_id, 'target': results['test']})
submission.to_csv('submission.csv', index=False)